Desert transcriptomics: assessing the genetic basis of adaptation to aridity in desert dwellers
Research areas: Research area(s): Ecology and Biodiversity of Plants and Ecosystems, Plant genetics
Principal investigators: Dr. Julia Bechteler, Prof. Dr. Dietmar Quandt, Prof. Dr. Thomas Wiehe
Project Info: Phase 3
To understand the various strategies that plants have evolved to adapt to arid habitats, information on diverse plant groups beside the standard model systems (e.g., Arabidopsis) is needed. We focus on the Loasaceae as study system, which enables comparative analyses due to several evolutionary transitions between habitats (arid-mesic-humid) and the independent colonization of two arid deserts, the Atacama and Namib. In order to identify adaptive genetic variation that evolved under similar selective pressure (i.e., similar environments) in arid habitats, we will compare between Huidobria fruticosa and H. chilensis, two perennial shrubs from the Loasaceae that are characteristic for the Quebrada systems in the Atacama Desert. Two environmentally different transects per species will be established in the Atacama to collect genotype data (B01) and ecological information (B01/B07) on the two species, which will be tested in RNASeq experiments under controlled conditions. We will complement our transcriptome analyses with physiological measurements, and a diverse set of environmental data, such as climate variables (A-projects) and substrate and leaf element composition (C-projects). In our gene expression analyses, we will focus on genes involved in osmotic stress responses, element (ion) uptake and transport, here specifically on genes related to the accumulation of the element (Si). Silicon, taken up by plants as silicic acid (Si(OH)4), is known to influence the abiotic stress tolerance in plants and it is incorporated in the trichomes of the Loasaceae species in focus. We will validate our results from the gene expression analyses by performing genome-wide-association studies (GWAS) in one of the two Huidobria species.
Phase 2
Desert transcriptomics: assessing the genetic basis of adaptation to aridity in desert dwellers
Research areas: Plant genetics, plant biochemistry and biophysics, evolution and systematics of plants and fungi
Principal investigators: Dr. Julia Bechteler, Prof. Dr. Dietmar Quandt, Prof. Dr. Thomas Wiehe
 |
|
|
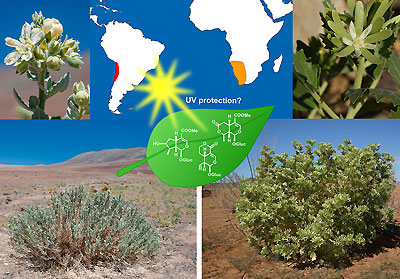
Loasaceae desert dwellers sharing iridoid secondary compounds. |
Desert environments, such as the Atacama or the Namib, with their extreme environmental conditions set high demands on its inhabiting species. Plants have adapted to their various stress factors, however, little is known about the genetic basis of such adaptations that is of importance in the evolution of desert plants. Especially the lack of comparative studies still precludes generalizations. Similarly, knowledge about iridoids, secondary compounds that are thought to play a role in herbivore deterrence and according to our preliminary results may act as sun protection in desert plants is still limited. Using comparative transcriptomics, and integrating biological, chemical, and geological data, we want to address our central question: “is there a common gene set which is relevant for adaptation to aridity in desert dwellers?”
We will compare the transcriptomes of six Loasaceae desert dwellers from the Atacama (Huidobria fruticosa, Presliophytum species) and Namib (Kissenia capensis) with those of their mesic relatives. We will test for signatures of natural selection in water stress responsive genes in the desert species. Gene expression studies with Huidobria fruticosa under controlled conditions in culture and at two climatically different sites in the Atacama Desert will provide insights into the ‘stress anticipation’ ability of desert plants – an important factor in the adaptation to extreme habitats.
Biochemical studies will complement our transcriptomic analyses. We will use Huidobria fruticosa, Kissenia capensis, and their mesic counterpart Xylopodia klaprothioides to identify iridoid compounds with state-of-the-art secondary compound analyses. We will use the generated transcriptomes to investigate the iridoid biosynthesis pathway, the underlying genes and gene regulatory mechanisms in the targeted Loasaceae species.
Publications
Project B7 - Publications
Articles
Hakobyan, A., Velte, S., Sickel, W., Quandt, D., Stoll, A., Knief, C., 2023.
Tillandsia landbeckii phyllosphere and laimosphere as refugia for bacterial life in a hyperarid desert environment.
Microbiome. 11 (246), 1 - 18. DOI: https://doi.org/10.1186/s40168-023-01684-x.
Schmitz, J., Bechteler, J., Münker, C., 2023.
Leaf element composition of the desert shrub Huidobria chilensis (Loasaceae): potential correlation with genetic structure, leaf age and substrate element composition.
Bachelor Thesis. 1 - 84.
Agnes, M., Bechteler, J., 2022.
Technical and biological aspects of whole-genome assembly and analysis of Huidobria chilensis and H. fruticosa (Loasaceae) from the Atacama Desert in Chile.
Bachelor Thesis. 1 - 72.
Iaboli, L., Bechteler, J., 2022.
Transcriptomic studies in seedlings of two different populations of the desert plant H. chilensis.
Master Thesis. 1 - 31.
Zilken, P., Bechteler, J., Quandt, D., 2022.
Evolutionary history of Copiapoa (Cactaceae) in the Atacama Desert based on a plastid marker dataset.
Bachelor Thesis. 1 - 47.
Event Paper
Morales Sanhueza, P. C., Iaboli, L., Ferchichi, K., Hüttel, B., Stoll, A., Agnes, M., Wiehe, T., Quandt, D., Bechteler, J., 2022.
Genetic basis of plant adaptation to aridity through evolutionary and ecological transcriptomics and genomics.
Proc. of Botanik Tagung 2022, August 29 - September 01, 2022, Main building of the University of Bonn , Regina-Pacis-Weg 3, 146 - 146.
Data
Project B7 - Research Data
Morales Sanhueza, P., Bechteler, J., 2024.
RNA raw sequences Huidobria chilensis populations in the Atacama Desert.
CRC1211 Database (CRC1211DB).
Morales Sanhueza, P., Bechteler, J., 2024.
Whole Genome Sequencing raw data for Loasaceae family species.
CRC1211 Database (CRC1211DB).
Morales Sanhueza, P., Bechteler, J., 2024.
RNA raw sequences Loasaceae family species.
CRC1211 Database (CRC1211DB).




























